Sep 14, 2022
Reminder: Links to course materials and main sites (Piazza, Canvas, Github) can be found on the home page of the main course website:
https://musa-550-fall-2022.github.io/
Week #2 repository: https://github.com/MUSA-550-Fall-2022/week-2
Recommended readings for the week listed here
Last time
Today
# The imports
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
So many tools...so little time
You'll use different packages to achieve different goals, and they each have different things they are good at.
Today, we'll focus on:
And next week for geospatial data:
Goal: introduce you to the most common tools and enable you to know the best package for the job in the future
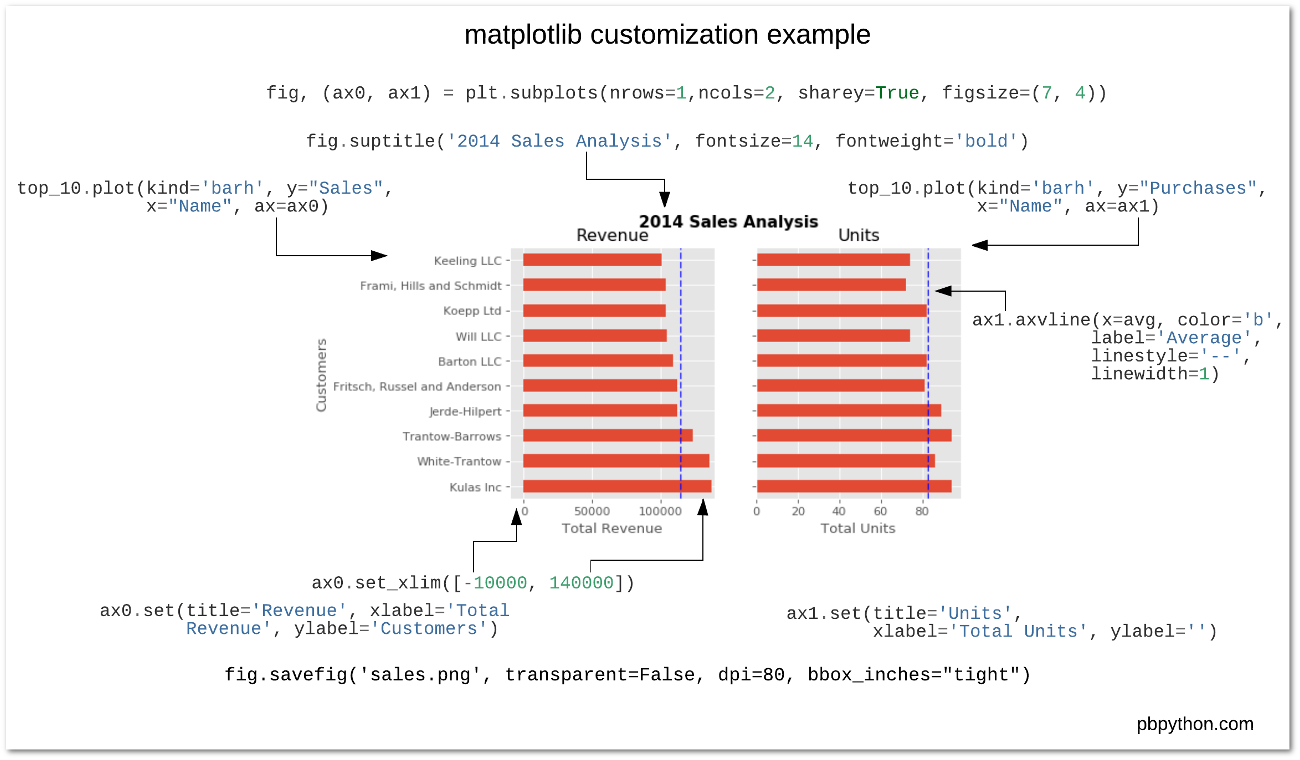
We'll use the object-oriented interface to matplotlib
Create Figure and Axes objects
Add plots to the Axes object
Customize any and all aspects of the Figure or Axes objects
Pro: Matplotlib is extraordinarily general — you can do pretty much anything with it
Con: There's a steep learning curve, with a lot of matplotlib-specific terms to learn
We'll use the Palmer penguins data set, data collected for three species of penguins at Palmer station in Antartica


Artwork by @allison_horst
# Load data on Palmer penguins
penguins = pd.read_csv("./data/penguins.csv")
penguins.head(n=10)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | male | 2007 |
| 6 | Adelie | Torgersen | 38.9 | 17.8 | 181.0 | 3625.0 | female | 2007 |
| 7 | Adelie | Torgersen | 39.2 | 19.6 | 195.0 | 4675.0 | male | 2007 |
| 8 | Adelie | Torgersen | 34.1 | 18.1 | 193.0 | 3475.0 | NaN | 2007 |
| 9 | Adelie | Torgersen | 42.0 | 20.2 | 190.0 | 4250.0 | NaN | 2007 |
Data is already in tidy format
I want to scatter flipper length vs. bill length, colored by the penguin species
# Initialize the figure and axes
fig, ax = plt.subplots(figsize=(10, 6))
# Color for each species
color_map = {"Adelie": "#1f77b4", "Gentoo": "#ff7f0e", "Chinstrap": "#D62728"}
# Group the data frame by species and loop over each group
# NOTE: "group" will be the dataframe holding the data for "species"
for species, group in penguins.groupby("species"):
print(f"Plotting {species}...")
# Plot flipper length vs bill length for this group
ax.scatter(
group["flipper_length_mm"],
group["bill_length_mm"],
marker="o",
label=species,
color=color_map[species],
alpha=0.75,
)
# Format the axes
ax.legend(loc="best")
ax.set_xlabel("Flipper Length (mm)")
ax.set_ylabel("Bill Length (mm)")
ax.grid(True)
# Show
plt.show()
Plotting Adelie... Plotting Chinstrap... Plotting Gentoo...
pandas?¶# Tab complete on the plot attribute of a dataframe to see the available functions
#penguins.plot.scatter?
# Initialize the figure and axes
fig, ax = plt.subplots(figsize=(10, 6))
# Calculate a list of colors
color_map = {"Adelie": "#1f77b4", "Gentoo": "#ff7f0e", "Chinstrap": "#D62728"}
colors = [color_map[species] for species in penguins["species"]]
# Scatter plot two columns, colored by third
penguins.plot.scatter(
x="flipper_length_mm",
y="bill_length_mm",
c=colors,
alpha=0.75,
ax=ax, # Plot on the axes object we created already!
)
# Format
ax.set_xlabel("Flipper Length (mm)")
ax.set_ylabel("Bill Length (mm)")
ax.grid(True)
Note: no easy way to get legend added to the plot in this case...
pandas plotting capabilities are good for quick and unpolished plots during the data exploration phaseimport seaborn as sns
# Initialize the figure and axes
fig, ax = plt.subplots(figsize=(10, 6))
# style keywords as dict
color_map = {"Adelie": "#1f77b4", "Gentoo": "#ff7f0e", "Chinstrap": "#D62728"}
style = dict(palette=color_map, s=60, edgecolor="none", alpha=0.75)
# use the scatterplot() function
sns.scatterplot(
x="flipper_length_mm", # the x column
y="bill_length_mm", # the y column
hue="species", # the third dimension (color)
data=penguins, # pass in the data
ax=ax, # plot on the axes object we made
**style # add our style keywords
)
# Format with matplotlib commands
ax.set_xlabel("Flipper Length (mm)")
ax.set_ylabel("Bill Length (mm)")
ax.grid(True)
ax.legend(loc='best')
<matplotlib.legend.Legend at 0x14398e0a0>
The ** syntax is the unpacking operator. It will unpack the dictionary and pass each keyword to the function.
So the previous code is the same as:
sns.scatterplot(
x="flipper_length_mm",
y="bill_length_mm",
hue="species",
data=penguins,
ax=ax,
palette=color_map, # defined in the style dict
edgecolor="none", # defined in the style dict
alpha=0.5 # defined in the style dict
)
But we can use **style as a shortcut!
In general, seaborn is fantastic for visualizing relationships between variables in a more quantitative way
Don't memorize every function...
I always look at the beautiful Example Gallery for ideas.
How about adding linear regression lines?
Use lmplot()
sns.lmplot(
x="flipper_length_mm",
y="bill_length_mm",
hue="species",
data=penguins,
height=6,
aspect=1.5,
palette=color_map,
scatter_kws=dict(edgecolor="none", alpha=0.5),
);
Use jointplot()
sns.jointplot(
x="flipper_length_mm",
y="bill_length_mm",
data=penguins,
height=8,
kind="kde",
cmap="viridis",
);
Use pairplot()
# The variables to plot
variables = [
"species",
"bill_length_mm",
"flipper_length_mm",
"body_mass_g",
"bill_depth_mm",
]
# Set the seaborn style
sns.set_context("notebook", font_scale=1.5)
# make the pair plot
sns.pairplot(
penguins[variables].dropna(),
palette=color_map,
hue="species",
plot_kws=dict(alpha=0.5, edgecolor="none"),
)
<seaborn.axisgrid.PairGrid at 0x143bf3eb0>
sns.catplot(x="species", y="bill_length_mm", hue="sex", data=penguins);
Great tutorial available in the seaborn documentation
The color_palette function in seaborn is very useful. Easiest way to get a list of hex strings for a specific color map.
viridis = sns.color_palette("viridis", n_colors=7).as_hex()
print(viridis)
['#472d7b', '#3b528b', '#2c728e', '#21918c', '#28ae80', '#5ec962', '#addc30']
sns.palplot(viridis)
You can also create custom light, dark, or diverging color maps, based on the desired hues at either end of the color map.
sns.palplot(sns.diverging_palette(10, 220, sep=50, n=7))
import altair as alt
Important: focuses on tidy data — you'll often find yourself running pd.melt() to get to tidy format
Let's try out our flipper length vs bill length example from last lecture...
# initialize the chart with the data
chart = alt.Chart(penguins)
# define what kind of marks to use
chart = chart.mark_circle(size=60)
# encode the visual channels
chart = chart.encode(
x="flipper_length_mm",
y="bill_length_mm",
color="species",
tooltip=["species", "flipper_length_mm", "bill_length_mm", "island", "sex"],
)
# make the chart interactive
chart.interactive()
Example: previous code is the same as
chart = chart.encode(
x=alt.X("flipper_length_mm"),
y=alt.Y("bill_length_mm"),
color=alt.Color("species"),
tooltip=alt.Tooltip(["species", "flipper_length_mm", "bill_length_mm", "island", "sex"]),
)
alt.Scale() object to specify the scale# initialize the chart with the data
chart = alt.Chart(penguins)
# define what kind of marks to use
chart = chart.mark_circle(size=60)
# encode the visual channels
chart = chart.encode(
x=alt.X("flipper_length_mm", scale=alt.Scale(zero=False)),
y=alt.Y("bill_length_mm", scale=alt.Scale(zero=False)),
color="species",
tooltip=["species", "flipper_length_mm", "bill_length_mm", "island", "sex"],
)
# make the chart interactive
chart = chart.interactive()
chart
For a complete list of these encodings, see the Encodings section of the documentation.
Altair charts can be fully specified as JSON $\rightarrow$ easy to embed in HTML on websites!
# Save the chart as a JSON string!
json = chart.to_json()
# Print out the first 1,000 characters
print(json[:1000])
{
"$schema": "https://vega.github.io/schema/vega-lite/v4.17.0.json",
"config": {
"view": {
"continuousHeight": 300,
"continuousWidth": 400
}
},
"data": {
"name": "data-d00e1631cca48c544438d30d2b470e8a"
},
"datasets": {
"data-d00e1631cca48c544438d30d2b470e8a": [
{
"bill_depth_mm": 18.7,
"bill_length_mm": 39.1,
"body_mass_g": 3750.0,
"flipper_length_mm": 181.0,
"island": "Torgersen",
"sex": "male",
"species": "Adelie",
"year": 2007
},
{
"bill_depth_mm": 17.4,
"bill_length_mm": 39.5,
"body_mass_g": 3800.0,
"flipper_length_mm": 186.0,
"island": "Torgersen",
"sex": "female",
"species": "Adelie",
"year": 2007
},
{
"bill_depth_mm": 18.0,
"bill_length_mm": 40.3,
"body_mass_g": 3250.0,
"flipper_length_mm": 195.0,
"island": "Torgersen",
"sex": "female",
chart.save("chart.html")
# Display IFrame in IPython
from IPython.display import IFrame
IFrame('chart.html', width=600, height=375)
chart = (
alt.Chart(penguins)
.mark_circle(size=60)
.encode(
x=alt.X("flipper_length_mm", scale=alt.Scale(zero=False)),
y=alt.Y("bill_length_mm", scale=alt.Scale(zero=False)),
color="species:N",
)
.interactive()
)
chart
Note that the interactive() call allows users to pan and zoom.
Altair is able to automatically determine the type of the variable using built-in heuristics. Altair and Vega-Lite support four primitive data types:
| Data Type | Code | Description |
|---|---|---|
| quantitative | Q | Numerical quantity (real-valued) |
| nominal | N | Name / Unordered categorical |
| ordinal | O | Ordered categorial |
| temporal | T | Date/time |
You can set the data type of a column explicitly using a one letter code attached to the column name with a colon:
Easily create multiple views of a dataset.
(
alt.Chart(penguins)
.mark_point()
.encode(
x=alt.X("flipper_length_mm:Q", scale=alt.Scale(zero=False)),
y=alt.Y("bill_length_mm:Q", scale=alt.Scale(zero=False)),
color="species:N"
).properties(
width=200, height=200
).facet(column="species").interactive()
)
Note: I've added the variable type identifiers (Q, N) to the previous example
Lots of features to create compound charts: repeated charts, faceted charts, vertical and horizontal stacking of subplots.
See the documentation for examples
A relatively new addition to altair, vega, and vega-lite. This allows you to define what happens when users interact with your visualization.
# create the selection box
brush = alt.selection_interval()
alt.Chart(penguins).mark_point().encode(
x=alt.X(
"flipper_length_mm", scale=alt.Scale(zero=False)
), # x
y=alt.Y(
"bill_length_mm", scale=alt.Scale(zero=False)
), # y
color=alt.condition(
brush, "species", alt.value("lightgray")
), # color
tooltip=["species", "flipper_length_mm", "bill_length_mm"],
).properties(
width=200, height=200, selection=brush
).facet(column="species")
We used the alt.condition() function to specify a conditional color for the markers. It takes three arguments:
brush object determines if abrush, color the marker according to the "species" columnbrush, use the literal hex color "lightgray"Let's examine the relationship between flipper_length_mm, bill_length_mm, and body_mass_g
We'll use a repeated chart that repeats variables across rows and columns.
Use a conditional color again, based on a brush selection.
# Setup the selection brush
brush = alt.selection(type='interval', resolve='global')
# Setup the chart
alt.Chart(penguins).mark_circle().encode(
x=alt.X(alt.repeat("column"), type='quantitative', scale=alt.Scale(zero=False)),
y=alt.Y(alt.repeat("row"), type='quantitative', scale=alt.Scale(zero=False)),
color=alt.condition(brush, 'species:N', alt.value('lightgray')), # conditional color
).properties(
width=200,
height=200,
selection=brush
).repeat( # repeat variables across rows and columns
row=['flipper_length_mm', 'bill_length_mm', 'body_mass_g'],
column=['body_mass_g', 'bill_length_mm', 'flipper_length_mm']
)
Let's explore the relationship between flipper length, body mass, and sex.
Scatter flipper length vs body mass for each species, colored by sex
alt.Chart(penguins).mark_point().encode(
x=alt.X('flipper_length_mm', scale=alt.Scale(zero=False)),
y=alt.Y('body_mass_g', scale=alt.Scale(zero=False)),
color=alt.Color("sex:N", scale=alt.Scale(scheme="Set2")),
).properties(
width=400, height=150
).facet(row='species')
I've specified the scale keyword to the alt.Color() object and passed a scheme value:
scale=alt.Scale(scheme="Set2")
Set2 is a Color Brewer color. The available color schemes are very similar to those matplotlib. A list is available on the Vega documentation: https://vega.github.io/vega/docs/schemes/.
Next, plot the total number of penguins per species by the island they are found on.
(
alt.Chart(penguins)
.mark_bar()
.encode(
x=alt.X('*:Q', aggregate='count', stack='normalize'),
y='island:N',
color='species:N',
tooltip=['island','species', 'count(*):Q']
)
)
Plot a histogram of number of penguins by flipper length, grouped by species.
(
alt.Chart(penguins)
.mark_bar()
.encode(
x=alt.X('flipper_length_mm', bin=alt.Bin(maxbins=20)),
y='count():Q', #shorthand
color='species',
tooltip=['species', alt.Tooltip('count()', title='Number of Penguins')]
).properties(height=250)
)
Finally, let's bin the data by body mass and plot the average flipper length per bin, colored by the species.
(
alt.Chart(penguins.dropna())
.mark_line()
.encode(
x=alt.X("body_mass_g:Q", bin=alt.Bin(maxbins=10)),
y=alt.Y('mean(flipper_length_mm):Q', scale=alt.Scale(zero=False)), # apply a mean to the flipper length in each bin
color='species:N',
tooltip=['mean(flipper_length_mm):Q', "count():Q"]
).properties(height=300, width=500)
)
In addition to mean() and count(), you can apply a number of different transformations to the data before plotting, including binning, arbitrary functions, and filters.
See the Data Transformations section of the user guide for more details.
# Setup a brush selection
brush = alt.selection(type='interval')
# The top scatterplot: flipper length vs bill length
points = (
alt.Chart()
.mark_point()
.encode(
x=alt.X('flipper_length_mm:Q', scale=alt.Scale(zero=False)),
y=alt.Y('bill_length_mm:Q', scale=alt.Scale(zero=False)),
color=alt.condition(brush, 'species:N', alt.value('lightgray'))
).properties(
selection=brush,
width=800
)
)
# the bottom bar plot
bars = (
alt.Chart()
.mark_bar()
.encode(
x='count(species):Q',
y='species:N',
color='species:N',
).transform_filter(
brush.ref() # the filter transform uses the selection to filter the input data to this chart
).properties(width=800)
)
chart = alt.vconcat(points, bars, data=penguins) # vertical stacking
chart
Exercise: let's reproduce this famous Wall Street Journal visualization showing measles incidence over time.
http://graphics.wsj.com/infectious-diseases-and-vaccines/